Acknowledgments: An earlier version of the paper, Linking Everything to Everything: Journal Publishing Myth or Reality?, was first presented at the ICCC/IFIP Conference on Electronic Publishing ‘97: New Models and Opportunities held in Canterbury, UK, in April 1997.
This latest version is revised from the original, containing updates and revisions in response to issues raised by the editors of Serials Review. We thank them for their input, and for allowing us to reproduce the work here.
PDF linking applications were developed by Professor David Brailsford,
Steve Probets and David Evans in the Electronic
Publishing Research Group at Nottingham University.
![]() †
The Open Journal project was funded by JISC's Electronic
Libraries (eLib) Programme award ELP2/35.
†
The Open Journal project was funded by JISC's Electronic
Libraries (eLib) Programme award ELP2/35. ![]()
Commercially-produced online journals are entering a new phase. In a little over a year those publishers that were among the first to make substantial journal programmes available online have begun to add features which are not directly available in corresponding print editions. The agenda has moved on from how you put journals online to how those journals can be enhanced.
Prior to the wider availability of online journals, talk of enhancements tended to focus on the idea of 'multimedia' content. Although some fields such as medicine and biology may be in a position to build and use such materials, the widespread realisation of audio-visual support for essentially text-based journals, as well as an adequate network infrastructure to distribute such materials, is still some way off.
Instead, attention has focussed on the hypertext link. In fact, links are a vital component of integrating multimedia content created in widely differing formats, but as far as online journals are concerned the first application of links on a large scale extends a convention that is fundamental to the modern academic journal: the use of citations.
Through reference lists within primary journal articles and the proliferation of secondary information sources - indexing and abstracting services, reviews, etc. - the journal literature is intrinsically 'hyperlinked', but the online medium is a more natural environment for this feature. The essence of the online environment is speed of access to the linked materials (Hitchcock 1996). In principle electronic links can be followed in an instant and will prove to be orders of magnitude more productive for the user than hyperlinks in print. In the electronic domain links can give the user access to a cited resource, possibly in its full form or to further information about that resource. Online journals that demonstrate citation linking are appearing in the areas of biology, physics and astronomy (Hitchcock et al. 1997).
This paper will examine two approaches being used to create
links - citation links, but also authored links to non-traditional sources
such as databases and other reference sources. The objective in both cases
is to create and maintain large numbers of links, potentially linking everything
to everything. One approach is that currently used by Electronic Press
Ltd, a commercial publisher and producer of online journals, for the BioMedNet
online club for those working in biology and medicine. Also examined is
the Open Journal research project, which involves the novel approach of
storing the link information separately from the authored documents, thereby
potentially improving the flexibility of linking and creating a new, reusable
and possibly valuable resource. By comparing the two approaches it may
be possible to identify which features will be important in providing the
most practical, efficient and cost-effective method for creating and maintaining
links, a service that will become a vital component of online publishing.
The Web has become a massively popular Internet service
in just 2-3 years since the introduction of graphical browsers such as
Mosaic and Netscape. During these early years much of the content of the
Web has been text-based, and the single dominating feature of text on the
Web is the ‘blue button’ link. So it may be possible to conclude, not only
intuitively as did Bush (1945), that links between
properly connected pieces of information are important. The popularity
of the Web shows there is a real demand for links when they are simple
to create and to use.
In the latter case it is the publishing framework that finds or creates value in something that may not inherently contain that value outside the framework. Through services such as BioMedNet (Quek and Tarr 1996), this framework is beginning to develop on the Web. The BioMedNet Library contains 250 full-text publications and a further 200 will be added in 1998. The club has 180,000 active users (growing at a rate of more than 2000 per week). Club members in mid-1997 were accessing 320,000 pages a week, and have downloaded more than a quarter of a million full-text articles.
According to Tim O’Reilly, books and online publisher, early online products have added searchability and multimedia: ‘But the Web shows a third key advantage of an online product: the creation of information interfaces’. (O’Reilly 1996) From the user perspective, the need will be to be able to find information on demand quickly, accurately and reliably, whatever the data type (audio, video, graphical, etc.), often starting from an imperfectly formed query. The challenge for online publishers seeking to add value to Web content is to develop interfaces that will locate and deliver the requested information based on the optimum data type.
In this scenario links, and the distinction with which
links are applied, will become one of the principal determinants in establishing
the competitive position of the information seller.
EP’s goal of linking everything to everything, as well as providing bidirectional linking, can only be achieved if a given document links to all other documents that contain relevant information. For the BioMedNet library a level of relevance has, for practical reasons, been defined as all referring, or referred to, documents. To achieve bidirectional linking, there must be control over both source and destination of links to provide back links. In a commercial environment, however, there are restrictions preventing this. Data suppliers do not allow modifications to their databases yet users expect value to be added by data providers.
This section presents an overview of EP’s linking system
(‘Clinky’) and the external link database (‘BundledLinks’) that facilitates
commercial competition with database suppliers while maintaining scalability,
and discusses how EP achieves its goals with this implementation.
EP’s approach to Medline linking on a commercial scale, involving linking of at least 500k documents from a 16 GB of data, is based on a number of requirements:
This approach discards the node/link hypertext model in favour of a set-based model of direct node intersections (Parunak 1991) or ‘complex relations’ in which the hypertext takes on many properties of a sophisticated database (Marshall et al. 1991).
There is little human involvement in creating the links, which are generated programmatically by querying the bibliographic database with data from SGML tagged references. This query is similar to the common notion of a Standard or Structured Query Language (SQL) used in relational databases. The query is constructed from the last name of the first-named author, the longest word in the title, the volume, issue and start page numbers, the date of publication and a ‘mangled’ journal identifier. Generation of standard matching keys can also be used. An example of this is the use of standard Serial Item and Contribution Identifier (SICI) codes for matching articles published in journals or, in the case of Medline-aware documents, the use of Medline accession numbers.
The 'mangling' process involves looking up the cited journal title in a list of journals known to be in Medline (compiled from a published list of abbreviated names for journals called List of Journals Published in Index Medicus), and converting it into the three-letter Medline journal code. If a code exists for the reference in question, then there should be a Medline record to this reference. In practice there will be references that do not belong to the Medline set (such as theses, references to non-medical journal articles), and on average only about 60% of references in a typical medical paper are contained in the Medline data.
Of the references that can be found in Medline, it is still impossible to achieve complete linking. Both ends of the link are human typed and error prone: Medline records are input at the NLM, and the references by, or on behalf of, the author(s). In addition, errors can be introduced in the tagging process, either manually or programmatically.
The linking program is therefore forced to make a 'guesstimate' based on the available information, and this can achieve a success rate higher than 85%. For EP, given the scale of the database processing task, this is an economical and acceptable threshold. Currently EP only performs a single attempt at a match. If this fails, there is no attempt to resubmit the reference. A log of which records do not match and the reasons are made available so that, in theory, someone could go through the logs and hand-enter the mismatches, but this is not economical (Figures 1 and 2).
Stopped at : Sat Feb 01 19:21:31 1997
Those references for which the linking program can find matches can generate more than one match. The query generated for each reference will return a set of results ranked by relevance based on the seven information fields identified in the reference. The most relevant, i.e. the ‘best fit’, record is chosen so long as no less than five of the seven fields match and there is no more than one record with the highest ranking.
Another type of linking EP performs is to embed Medline
accession numbers (unique identification numbers) into the full-text journal
articles. This type of linking implies that by using the same linking mechanism,
Medline links can potentially be derived from any Medline database by constructing
a query based on the accession number. By storing only the accession number
and not a URL the link can be generated by a program, allowing the destination
to be changed ‘under’ the link. One problem with embedding accession numbers
is that as the database grows (e.g. adding Embase, Analytical Abstracts)
the number of accession numbers needed grows.
For example, once a biological research paper has been published on paper or in electronic form, it can always be referred to by its details or an address. If the same paper is then converted to an HTML document and is made available via the Web, it could take on a second address. The same paper might be abstracted for Medline as well as other similar database services such as Embase and Current Contents. Each database publisher may choose to augment the original reference with keywords, evaluations or abstractions, e.g. MeSH terms. The original publisher may choose to hold many different representations of the same document. Many documents published by the Current Science Group appear in BioMedNet as HTML and PDF documents.
For these reasons, within the BioMedNet Library some documents may be represented in four or more different ways, each of which is published by a different publisher which enforces its own rules on augmenting its own data. Each of the links is stored as a record akin to a database record, and are collected and stored in a bundle. A bundle is effectively a collection of links of representations of the same document. Each record can be thought of as an instance of an information unit, that unit being the sum of all its parts. It is not enough to think of the full-text article as the parent of these records as there may be more information held in annotations and abstracts of the original document than in the original document (Figures 3 and 4).
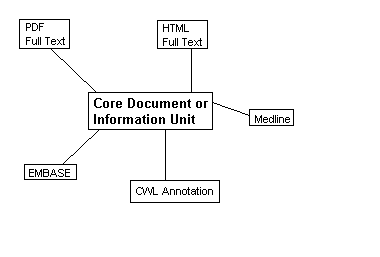
In many cases the bundle can be generated using data that
appears in the records. Many commercial databases store references to the
corresponding record in other databases. Medline accession numbers are
often used for this purpose. Utilising data directly from the records shifts
the linking burden to the database owner. Once two records from different
databases are identified as being instances of the same information unit,
it becomes possible to use either database identifier as a means of referring
to the bundle.
By collecting the instances of the record into a bundle then for each search a user performs the information units found can be identified and presented to the user with summary information about that unit instead of the usual "found records" display. In effect, if the user searches for a MeSH heading and finds a Medline record, the full-text version and user annotations can be indicated, where they exist, with very little overhead.
By using the bundle as a general storage area for holding meta-details of the information unit, these details can be made available as annotations to whatever document instance the user is viewing. If a user is viewing an abstract from an abstracting service they can also be provided with links to citations.
When a record is displayed in a Web browser, it becomes possible to provide details of which other representations are also available (Figure 5). A link to the PDF version of the full-text can be provided for printing.
The bundle is itself a record of the BundledLinks. In this way, not only are the links made available to all instances, but the problem of keeping all records synchronised is reduced to keeping a single entity synchronised. By representing documents as a source / identifier pair it is possible to change the data supplier without changing the destination of the link. So EP could, for example, for financial reasons choose to change document supplier for Medline without having to alter all links in the documents or in the BundledLinks. The hypertext link is generated on-the-fly from the bundle much as an HTML document is generated on-the-fly from the SGML document. For documents that exist only on the Web, the source can be represented as WWW and the identifier becomes the URL of the document.
With this structure it is possible to extract all the
links regardless of the record they actually appear in and present them
to the user regardless of the record they are viewing.
Being able to change the way the link is resolved (i.e. to change database provider) has commercial advantage not only for BioMedNet but users could configure their preferred data provider to avoid having to pay a new provider for data they already get from their own supplier. There are many ‘free’ Medline suppliers on the Web. Users should be able to choose which they prefer.
The commercial value of a link database was recognised before the Web. The Science Citation Index is an example of such a database. A bundle representing a research paper could be used either electronically or on paper for the same purposes.
EP’s internalised Medline linking may not be the best
approach for everyone, especially since there are now Medline services
on the Web, but it has enabled EP to achieve its self-imposed requirements
highlighted above. Part of the cost of innovation is that the solution
sought may not be there initially. Consequently, EP has a history of building
its own technologies such as Evaluated Medline. The important point, however,
is that the conceptual model must be workable, flexible and extensible,
and the linking mechanism is designed so that it does not depend on a closed
system to support external linking. The plan is to be able to implement
the same linking mechanism with other large-scale databases such as Embase.
In addition, EP is now seeking to extend its linking framework by participating
in the Open Journal project which is also applying link publishing software
as described below..
As the EP example shows, creating many thousands of links to support users in a specific knowledge domain is going to be a significant part of any works published online. Courseware developers have also discovered that creating and maintaining high-quality links is a major economic decision and not an after-thought at the end of the publishing process.
In contrast to EP’s internalised linking, one way of widening the use of links is to extend the way in which the Web itself supports the creation and implementation of links. Typically, Web links have to be authored within the source code of the original document and the link type used is limited; that is, Web links can simply point from one document to another. The key to exploiting the Web from a publishing perspective is to extend the Web as an open environment which supports linking not just as an authoring activity, but as a publishing task. In this respect linking becomes part of the value-adding process, where very large numbers of up-to-date links need to be added to every document to which they apply at the moment they are requested by the user. This could not be done by authors of individual papers.
More powerful ways of including links in documents are
being developed. The use of link services, linkbases and generic links
in the Open Journal model (Hitchcock et al. 1997)
are one way in which this can be achieved. Putting this in context requires
some understanding of the philosophy underlying the Web and of open hypertext,
or hypermedia, systems.
In this sense the Web is a classic open system. With its standard protocol for information transfer and universal addresses, anything that can be displayed can be interconnected. Simply, if the relationship between two works changes, the information 'could smoothly reshape to represent the new state of knowledge' (Berners-Lee et al. 1994).
The Web though is not an open hypertext system. A generally accepted requirement of open hypertext systems is that they do not differentiate between authors and readers. (Malcolm 1991) Each should be offered the same set of functions, that is, a reader should have the same facility for altering a version of a text, say, as the original author. By encoding links within HTML markup, the Web does not conform to this view, because it reduces linking to an author-only task. According to Berners-Lee et al. (1994): 'The Web does not yet meet its design goal as being a pool of knowledge that is as easy to update as to read.'
For ‘readers’ read ‘publishers’, because publishers have a greater need to manage content, and on the Web this will not always be content that the publisher ‘owns’, exactly as seen above. O’Reilly (1996) recognised the fundamental shift in publishing that the Web motivates: ‘In the old model, the information product is a container. In the new model, it is a core. One bounds a body of content, the other centers it’. According to Fillmore (1993), a founder of Open Book Systems: 'The successful online publisher will most likely license access to other people's content to supplement or enhance his own, whether that content is online books, databases, bulletin boards, graphic image repositories, or online advice columns. What's ‘for sale’ might be the interactive links, the thought structure the publisher puts around the distributed content area.'
The implications are enormous, but from a technical viewpoint
open hypertext systems which give publishers and users the option to make
links as well as follow links from third-party materials on the
Web, for example, are now supported commercially.
A number of open hypermedia systems reported in the early 1990s adopted a link service approach, including Microcosm (Fountain et al. 1990), Hyper-G (Kappe et al. 1992), and Multicard (Rizk and Sauter 1992). Two of these systems, Microcosm and Hyper-G, are now commercialised. There are other examples of open hypermedia systems in research (Wiil and Leggett 1997), but current interest centres on extending these linking models to augment the Web, such as the Distributed Link Service (Carr et al. 1995) and HyperWave (Maurer 1996).
As the Web is increasingly used to display documents created in common applications such as word processors and spreadsheets, which may or may not support HTML and thus hypertext capabilities, or which may or may not have authored links, the potential for a link service on the Web becomes apparent. 'Without a link service, Web users can follow links from HTML documents or pictures into dead-end media such as spreadsheets, CAD documents or text; with a link service they can also follow links out of these media again' (Carr et al. 1995).
It may be obvious to state that the effectiveness of a link service is predicated on the effectiveness of the links that it serves. The links served by the Distributed Link Service (DLS) and its commercial version Webcosm have semantics that are derived from the Microcosm system (Fountain et al. 1990). These allow a link to be parametrised against the identity of a document, against the position of the anchor within a document and against the data contents that the anchor selects. For example, each link is a pattern that can be matched against many potential documents to instantiate an actual link between two actual documents, that is, an instance of a link that might be seen when certain parameters prevail but which otherwise might not appear at all.
Thus, each link has the following format, and states the existence of a link from a source to a destination.
Both the source and destination are described as a triple: the document URL, the offset within the document, and the selected object within the document. The system pinpoints the link anchors either by measuring from the beginning of a document (using the offset), or by matching a selection, or both.
Links are of the following types:
Note how the DLS provides flexibility in specifying the source anchor: this means that a single link to a destination may appear in many places at once.
Links supported by the Web are specific, or ‘button’,
links. Each link has to be individually authored.
The projectis funded for three years to mid-1998 by the UK Electronic Libraries (eLib) research programme. and is among fifty or so projects within the programme, which aims to contribute to the international efforts to build integrated digital libraries. Evaluation is one of the features emphasized within eLib, both as a way of bringing the research into the community and to inform possible collaboration between different projects or even between projects in overseas programmes.
The project is developing three Open Journals in the areas of biology, cognitive science and computer science. Since the cultures and practices of each field tend to be reflected in the respective literatures, these in turn determine linking strategies. The characteristics of each Open Journal are already markedly different. The important feature here is how the system used for creating links adapts to the different requirements and copes with the formats in which the original materials are presented, the principal formats in this case being those popular for online journals, HTML and PDF. Link inclusion, from a linkbase, in PDF documents is supported in the project by applications developed in the Electronic Publishing Research Group (EPRG) at Nottingham University, and is an extension of that group's CAJUN (CD-ROM Acrobat Journals Using Networks) project (Smith et al. 1993).
The spectrum of link creation options supported by the DLS includes highly pertinent, hand-crafted links such as might take a user from a biology journal page to a graphical molecular database. Links can also be created en masse by a batch computational process, for example, in citation linking or linking complex terms to a definition in a specialised dictionary.
On the Web each link must be individually identified and specified, making it a cumbersome process especially if, as today, the navigation facilities of the native Web environment are not particularly advanced and do not aid the link creator sufficiently in browsing relevant resources. (O’Leary 1997) For this reason, the majority of the 12,000 links that are currently being demonstrated in the Biology Open Journal were created by computational methods and demonstrate the power of the generic link. A cheap way of providing a database of links for a journal archive is to create a link from any occurrence of a specific word or phrase within a literary corpus to any paper with that keyword specified. This approach can also be used for terms in an online dictionary, so that the occurrence of a key dictionary term anywhere in any document is automatically linked directly to its definition. (Figure 6) In fact, this is a variation on the generic link, where the document context is constrained to be ‘inside’ the boundaries of the Open Journal. So, any mention of the word ‘embryo’ may link to an entry in the online Dictionary of Cell Biology if it is found in the Biology Open Journal, but may not if the instance is inside, say, the Open Journal of Computing.
To create a database of these links requires a source of metadata for the articles of interest: extracting the keyword fields is a small programming effort which leverages links out of another’s authorial or editorial effort. In practice, many of the project’s resources are in PDF and have not been provided with metadata records, so the programming effort required to extract the keywords from an encoded document display format has been a lot higher - but still less than creating the links manually.
Although the project philosophy is to give users access
to links distributed across a network, it has found, as has EP with its
Medline linking, that access to localised, formattable data is necessary
for creating large linkbases computationally. The application of citation
linking in the Cognitive Science Open Journal is an example. Selected abstracts
data made available to the project by the Institute for Scientific Information
(ISI) required extensive reformatting, but the resulting links from journals
such as Psycoloquy are proving reliable and relatively complete
(Hitchcock et al. 1997). The real power of
this approach, however, is that once created, this referencing linkbase
could be applied to other cognitive science journals from which reference
data can be parsed wherever they are on the Web and wherever the abstracts
database is held. All that is required is that user is able to access the
resources, e.g. as a subscriber, as well as the link service.
Having created a set of links, the author can store them either in a single linkbase which must be chosen explicitly by an end-user, or amalgamated into a database of linkbases which can be chosen as part of a larger context. The former option allows for highly specific links, tailored for a small user population, but inevitably results in a large number of these collections. The latter option results in a more generally applicable database, but one that is of lesser relevance to any particular user.
As the project has developed, more of the linkbases have tended to be of the second type. Consequently, the DLS is being revised to give the user explicit control over the kinds of link that they would like to see. Links could be differentiated by colour (or simply pruned according to particular thresholds) according to whether they are hand-authored specific links, machine-generated general links, recently created links or links belonging to (for example) a course tutor. This adds to the user’s control over the view of the document’s connectivity, which currently exists only at the macro-level by including or excluding whole linkbases.
While the most successful strategies for link creation
have depended on data processed locally, the project aims to develop tools
to make more use of remote Web resources. One possible approach is to augment
an author’s resource discovery strategies by piggybacking an existing Web
search service. For example, an ActiveX-based interface could allow the
author to request that the results of keyword searches automatically be
turned into linkbases. This emulates the cheap link creation described
above, but also allows the user to view the results of the search from
a browser and prune the search results into the most pertinent set of keyword
links.
The application of links must be led, or constrained, by user expectations, although first impressions suggest these should not be underestimated, especially with regard to citation linking. Once established this feature is so powerful that it will be almost mandatory and will transform journal usage.
In other respects the ability to link everything to everything is not always desirable. The practice of citations is well established. In contrast the overlaying of keyword data as links on text is not, so first reactions to such links have been less positive. Simply, not enough is known about the effect on the literature for this type of linking to be applied indiscriminately on a large scale. A better understanding of when and where to use this approach allied to greater precision in the use of linkbases will make this approach more attractive if, in this case, the demand for universality of links is tempered.
EP’s approach enables it to represent what users expect from a commercial system while maintaining source databases as supplied by their owners. The data can change ‘underneath’ or ‘on-top-of’ the links without affecting the validity of the link. Effectively this is an open hypermedia approach, similar in principle to that being used to build Open Journals in biology and cognitive science.
What is now required in link publishing are tools such as the DLS to provide more explicit editorial control over the quality of links. By applying distributed open hypertext services such the DLS, we can begin to see that such an approach offers the flexibility and cost-effectiveness for large-scale link creation and maintenance that is not possible with the Web alone.
Most importantly perhaps, as EP shows, links stored in
bundles or linkbases have additional commercial value because the bundle
becomes a piece of information itself, quite distinct from the underlying
text. Exploiting this value is an area of investigation for the future.