Challenge
Start of topic | Skip to actions
Provenance Challenge Template
Submission in progressParticipating Team
- Short team name: RWS
- Participant names: Norbert Podhorszki, Ilkay Altintas, Shawn Bowers, Zhijie Guan, Bertram Ludaescher, Tim McPhillips.
- Project URL: http://kepler-project.org
- Project Overview: This small in-house project aims to use the RWS provenance model for nested Kepler workflows.
- Provenance-specific Overview: The current goal is to address single workflow runs and answer questions like the question1 but for nested workflows, for several iterations and for loops as well.
- Relevant Publications:
- A Model for User-Oriented Data Provenance in Pipelined Scientific Workflows, Shawn Bowers, Timothy McPhillips, Bertram Ludaescher, Shirley Cohen, Susan B. Davidson. International Provenance and Annotation Workshop (IPAW'06), Chicago, Illinois, USA, May 3-5, 2006.
- Provenance Collection Support in the Kepler Scientific Workflow System, Ilkay Altintas, Oscar Barney, Efrat Jaeger-Frank, International Provenance and Annotation Workshop (IPAW'06), Chicago, Illinois, USA, May 3-5, 2006.
Workflow Representation
Our workflow is represented in PtolemyII's MoML language, created in Kepler.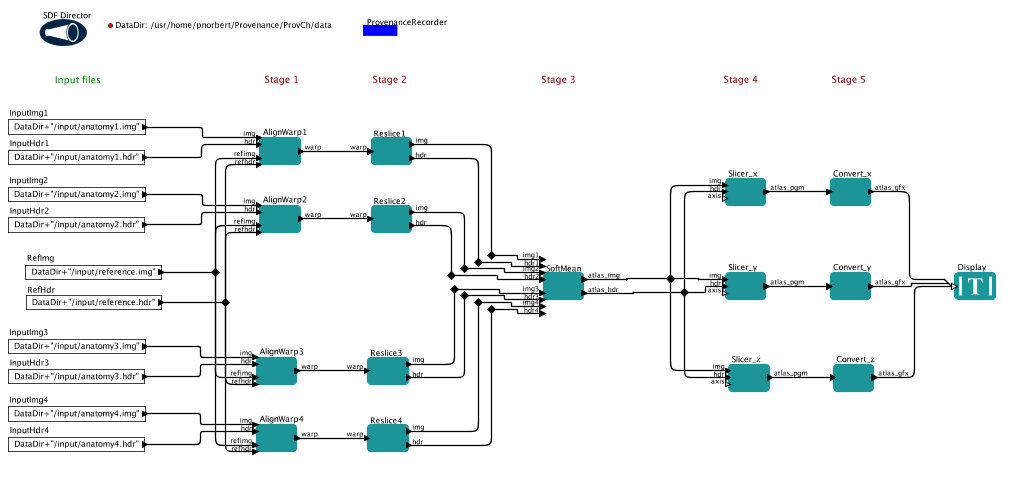
Figure 1. The Kepler workflow for the challenge
Notions:
- Actor
- The white and green boxes on screen are actors, that execute something.
- Port
- An actor have input/output ports on which it receives/sends tokens
- Token
- Data is embedded in tokens in the workflow. They are propagated in the workflow.
- SDF Director
- The computation model of this workflow is SDF (Synchronous Data Flow). This means sequential execution with a precomputed schedule (the most basic and simplest model of Kepler).
Provenance Trace
According to the RWS model, r(ead), w(rite) and s(tate-reset) event are recorded. Besides that we need to record the actors, their ports, the "tokens" flowing in the workflow among the actors, the created objects and their values. The RWS prototype inference engine is implemented in Prolog ;-), and the provenance data is currently printed out simply as Prolog fact set, but it will be put in relational database in the future.Tables (as Prolog predicates)
portTable Port - Actor relationship, and (atomic vs composite) type of port/actor
portTable('.pc.align_warp2.GetIndexFromName.StringIndexOf.output', '.pc.align_warp2.GetIndexFromName.StringIndexOf', a).
portTable('.pc.convert_x.atlas_gfx', '.pc.convert_x', c).
tokenTable Token - Object relationship. The object carries the data and it has a unique ID.
tokenTable('.pc.convert_x.atlas_gfx.0.0', o596169037_35902573).
objectTable Object - Value relationship. Currently no type is recorded.
objectTable(o596169037_35902573, '"/usr/home/pnorbert/Provenance/ProvCh/data/output/atlas-x.gif"', notype).
traceTable The RWS event trace, when a port is reading/writing a token, or an actor has a state-reset
traceTable('.pc.convert_x', s, 'nil', 1). - state reset of actor
traceTable('.pc.convert_x.input', r, '.pc.slicer_x.atlas_pgm.0.0', 1). - read a token on port 'input'
traceTable('.pc.convert_x.atlas_gfx', w, '.pc.convert_x.atlas_gfx.0.0', 1). - write token on port 'atlas_gfx'
Trace size of the flat challenge workflow:
| Table | lines |
|---|---|
| portTable | 81 |
| tokenTable | 30 |
| objectTable | 30 |
| traceTable | 86 |
Provenance Queries Matrix
| Teams | Queries | ||||||||||
| Q1 | Q2 | Q3 | Q4 | Q5 | Q6 | Q7 | Q8 | Q9 | |||
| RWS team |  |  |  |  |  |  |  |  |  | ||
Provenance Queries
The inference engine prototype is implemented in Prolog. The basic information we need is the token lineage of a given token, i.e. all tokens in the workflow on that the given token depended. Then values, objects and actors can be looked up from the provenance tables. The predicatetokenLineageOfValue( Value, List ) provides the list of
all tokens on that the given value (more accurately the token that
contained this value first) depended. This predicate basically
generates the transitive closure of the direct dependency among
tokens, back to the very first tokens, i.e. to the inputs.
valueLineageOfValue/2 and actorLineageOfValue/2 use the above
predicate and then look for the token's value it contains or the actor
that generated it, resp.
Since the dependency graph may contain several paths back to a certain
token and also several tokens can be created by the same actor, we may
get an actor or value in the list several times. Therefore, we use the
list_to_set/2 built-in predicate to make each resulted element
unique.
1. Find the process that led to Atlas X Graphic / everything that caused Atlas X Graphic to be as it is. This should tell us the new brain images from which the averaged atlas was generated, the warping performed etc.
Input: the output file name as "/usr/home/pnorbert/Provenance/ProvCh/data/output/atlas-x.gif" Output: a set of actors that contributed and data values (file names) that led to this file
Answer 1.a. List of actors that contributed to the result: (21 actors).
They appear in reversed order as they were executed.
?- q1_actors('"/usr/home/pnorbert/Provenance/ProvCh/data/output/atlas-x.gif"', ActorList), print(ActorList).
[
.pc.Convert_x,
.pc.Slicer_x,
.pc.SoftMean,
.pc.Reslice3,
.pc.Reslice2,
.pc.Reslice4,
.pc.Reslice1,
.pc.AlignWarp3,
.pc.RefImg,
.pc.RefHdr,
.pc.InputHdr3,
.pc.InputImg3,
.pc.AlignWarp2,
.pc.InputHdr2,
.pc.InputImg2,
.pc.AlignWarp4,
.pc.InputHdr4,
.pc.InputImg4,
.pc.AlignWarp1,
.pc.InputImg1,
.pc.InputHdr1
]
Note: new lines entered manually in the doc for easier read.
Answer b. List of input and intermediate values created by the workflow (26 values).
?- q1_values('"/usr/home/pnorbert/Provenance/ProvCh/data/output/atlas-x.gif"', ValueList), print(ValueList).
[
"/usr/home/pnorbert/Provenance/ProvCh/data/output/atlas-x.gif",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage4/atlas-x.pgm",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage3/atlas.img",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage3/atlas.hdr",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage2/resliced3.hdr",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage2/resliced2.img",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage2/resliced4.hdr",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage2/resliced1.img",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage2/resliced2.hdr",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage2/resliced3.img",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage2/resliced4.img",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage2/resliced1.hdr",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage1/warp3.warp",
"/usr/home/pnorbert/Provenance/ProvCh/data/input/reference.img",
"/usr/home/pnorbert/Provenance/ProvCh/data/input/reference.hdr",
"/usr/home/pnorbert/Provenance/ProvCh/data/input/anatomy3.hdr",
"/usr/home/pnorbert/Provenance/ProvCh/data/input/anatomy3.img",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage1/warp2.warp",
"/usr/home/pnorbert/Provenance/ProvCh/data/input/anatomy2.hdr",
"/usr/home/pnorbert/Provenance/ProvCh/data/input/anatomy2.img",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage1/warp4.warp",
"/usr/home/pnorbert/Provenance/ProvCh/data/input/anatomy4.hdr",
"/usr/home/pnorbert/Provenance/ProvCh/data/input/anatomy4.img",
"/usr/home/pnorbert/Provenance/ProvCh/data/out-stage1/warp1.warp",
"/usr/home/pnorbert/Provenance/ProvCh/data/input/anatomy1.img",
"/usr/home/pnorbert/Provenance/ProvCh/data/input/anatomy1.hdr"
]
Suggested Workflow Variants
Like the myGrid/Taverna team, we also created a more generic version of the challenge workflow, which works for any number of input images, provided that the Softmean can take any number of input images at once.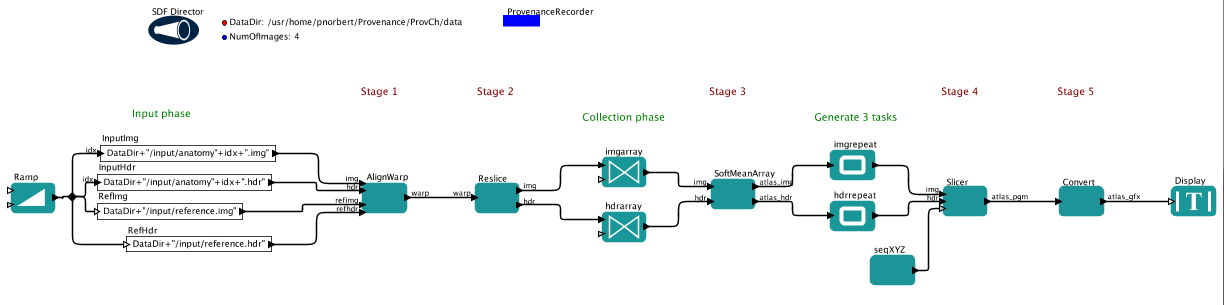
Figure 2. The generalized Kepler workflow for the challenge
The difference between this and the Taverna workflow is that we create the input file names within the workflow one-by-one. A list of input
files would be given as one token to the first AlignWrap actor, which supposedly wants to get them one-by-one. The 4 outputs of the Reslice are collected into an array and Softmean is executed only once. The output of Softmean is repeated 3 times with different slice parameters (generated by the seqXYZ actor), thus executing the final two operations three times.
The answers for the first query are basically the same but with some changes. The actor list is shorter, reporting e.g. Reslice once instead of Reslice1,...,Reslice4, however, the additional array and repeat operations appear in the list. The value list becomes larger because of the additional array and repetition tokens.
Suggested Queries
Categorisation of queries
Live systems
- Kepler: http://kepler-project.org
Further Comments
Conclusions
-- NorbertPodhorszki - 07 Sep 2006to top
| I | Attachment  | Action | Size | Date | Who | Comment |
|---|---|---|---|---|---|---|
| | pc.png | manage | 70.0 K | 07 Sep 2006 - 16:41 | NorbertPodhorszki | Kepler workflow, flat version |
| | pca.png | manage | 47.5 K | 07 Sep 2006 - 17:12 | NorbertPodhorszki | Generalized Kepler workflow |
| | trace-pc.txt | manage | 15.4 K | 07 Sep 2006 - 17:42 | NorbertPodhorszki | The provenance trace of the original workflow |
Edit | Attach image or document | Printable version | Raw text | More topic actions
Revisions: | r1.3 | > | r1.2 | > | r1.1 | Total page history | Backlinks
Revisions: | r1.3 | > | r1.2 | > | r1.1 | Total page history | Backlinks