WorkflowRepository
Submitted to Open Repositories 2009
The myExperiment Open Repository for Scientific Workflows
David De Roure (1), Carole Goble (2), Sergejs Aleksejevs (2), Sean Bechhofer (2), Jiten Bhagat (2), Don Cruickshank (1), Danius Michaelides (1), David Newman (1)
(1) Intelligence Agents Multimedia Group, School of Electronics and Computer Science, University of Southampton, UK
(2) Information Management Group, School of Computer Science, The University of Manchester, UK
ABSTRACT
myExperiment is an open repository solution for the born-digital items arising in contemporary research practice, in particular scientific workflows and experiment plans. Launched in November 2007, the public repository (myexperiment.org) has established a significant collection of scientific workflows, spanning multiple disciplines and multiple workflow systems, which has been accessed by over 16,000 users worldwide. Built according to Web 2.0 design principles, myExperiment demonstrates the success of blending modern social curation methods with the demands of researchers sharing hard-won intellectual assets and research works within a scholarly communication lifecycle. myExperiment is an important component in the revolution in creating, sharing and publishing scientific results, and has already established itself as a valuable and unique repository with a growing international presence.
Introduction
Researchers in every discipline now have access to a vast array of digital resources, ranging from libraries of articles and data collections to analytical tools and visualisation applications, many publicly available. These digital resources are combined and their data aggregated and analysed in the day-to-day conduct of research. With these new techniques come many new kinds of digital assets which have yet to become adopted within the scholarly knowledge lifecycle, yet they are an essential part of it. One of the most important is the scientific workflow. A scientific workflow is the description of a process that specifies the co-ordinated execution of multiple tasks so that, for example, data analysis and simulations can be repeated and accurately reported. Alongside experiment plans, Standard Operating Procedures and laboratory protocols, these automated workflows are one of the most recent forms of scientific digital methods, and one that has gained popularity and adoption in a short time [1]. They represent the methods component of modern research and are valuable and important scholarly assets in their own right.
Scientific workflows are valuable commodities which require expertise to build. The myExperiment repository was motivated by observing a clear need to share workflows – to reduce reinvention, propagate best practice and enable scientists to concentrate on science – amongst decoupled communities of workflow users. It was also motivated by a frustration with existing systems which: (a) missed the social dimension, merely making things available rather than encouraging and controlling sharing; (b) presented complex user interfaces out of line with the popular web sites that people are using on an everyday basis, thereby demanding further skill.
These methods are crucial intellectual assets of the research life cycle whose stewardship has been neglected. Repositories often emphasise curation of data, but in digital research the curation of the process around that data is equally important – hence by focusing on workflows, myExperiment also provides a mechanism for expert and community curation of process [2-3].
The myExperiment approach
The myExperiment open source software provides a repository for research objects, somewhat analogous to learning objects. A research object contains various digital research materials and methods – documents, simulations, data, scripts, workflows – coupled with a research objective or designed to support a research process. Research objects are intended to capture a scientific investigation or research question and are expected to be reusable, repeatable, and replayable.
Sharing research objects is a means to propagate practice and support reproducible research, but we believe it is not sufficient simply to make content available and assume a “build it and they will come” approach. Instead we have created an environment which facilitates the use of research objects and encourages sharing:
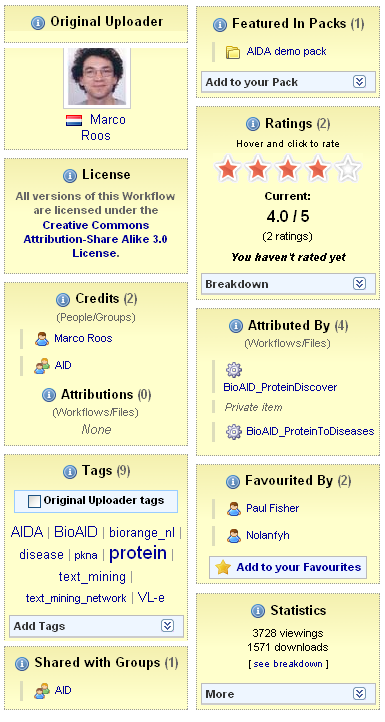
- Social sourcing and curation of content: myExperiment supports the social network of users and content, assisting researchers in discovering resources as well as in deposit, publication and curation. While it has parallels with websites such as FaceBook, myExperiment also provides a privacy, sharing, licensing, credit and attribution model which reflects and respects the needs of researchers and offers incentives for self-archiving and social curation. The “social metadata”, including tags and ratings, is shown in Figure 1.
- Familiar user experience: myExperiment provides an environment which is focused on the user experience, providing an attractive and immediately understandable web interface that uses the metaphors and behaviours of popular tools used in everyday life. It is immediately familiar to a new generation of students and researchers.
- Seamless access: Importantly, myExperiment provides a simple and powerful developer interface (REST API) that readily brings myExperiment’s functionality into the researcher’s own work environment so that it is seamlessly accessible at point of delivery, encouraging deposit and facilitating access. The API has enabled new interfaces to be built, such as Google Gadgets and Facebook Apps. It also enables existing interfaces to incorporate myExperiment functionality, such as a wiki, and the creation of functionality “mashups”.
Through its design to support Research Objects, myExperiment recognises that researchers work not just with individual items but with collections of items associated with a particular study. These may be stored locally, in multiple repositories or as native Web content. Hence research objects in myExperiment can be aggregations of items stored in the myExperiment repository as well as elsewhere. In myExperiment these are called “packs”, and while a pack might aggregate external content stored in multiple specialised repositories for particular content types, the pack itself is a single entity which can be tagged, reviewed, published, shared etc. For example, a pack might correspond to an experiment, containing input and output data, the experiment plan, associated publications and presentations, enabling that experiment to be shared; another example is a pack containing all the evidence corresponding to a particular decision as part of the record of the research process. Packs are described using the OAI’s Object Reuse and Exchange representation.
Like EPrints, the myExperiment software can be downloaded and installed locally, and it has basic federation capabilities. Significantly we also operate the public instance, myexperiment.org, providing a community focus which maximises users and content and nurtures network effects – as the collection grows and makes discovery and curation more difficult, the social network grows to facilitate both. myexperiment.org has gathered a significant user community worldwide and caught the imagination of the scientific and the Web communities with its vision that scientists should be able to swap workflows and other scientific objects as easily as citizens can share documents, photos and videos on the Web. Citations include New Scientist and Nature [4-6],
myExperiment is designed to call upon external services in order to process research objects. For example, scientific workflows are executed by myExperiment submitting a collection of research objects for remote processing to an enactor, and the results are automatically collected back into myExperiment. A similar mechanism could run simulations, scripts or statistical models. Similarly, workflows can be executed over myExperiment services for purposes of search or curation. Hence myExperiment serves as an example of integrating digital library content with computational tools and services, and also as an illustration of the role of workflows in the digital library.
These repository innovations – the social website, sharing of objects and process, support for aggregations, a unique public collection and the ability to “action” research – are documented in our publications in the international scientific community [7], as is our sociotechnical approach to the design [8] and also the way in which it has been delivered [9]. The site and the software have become the focus of an increasing number of collaborations in the publishing, digital libraries and scholarly publishing communities.
Building myExperiment
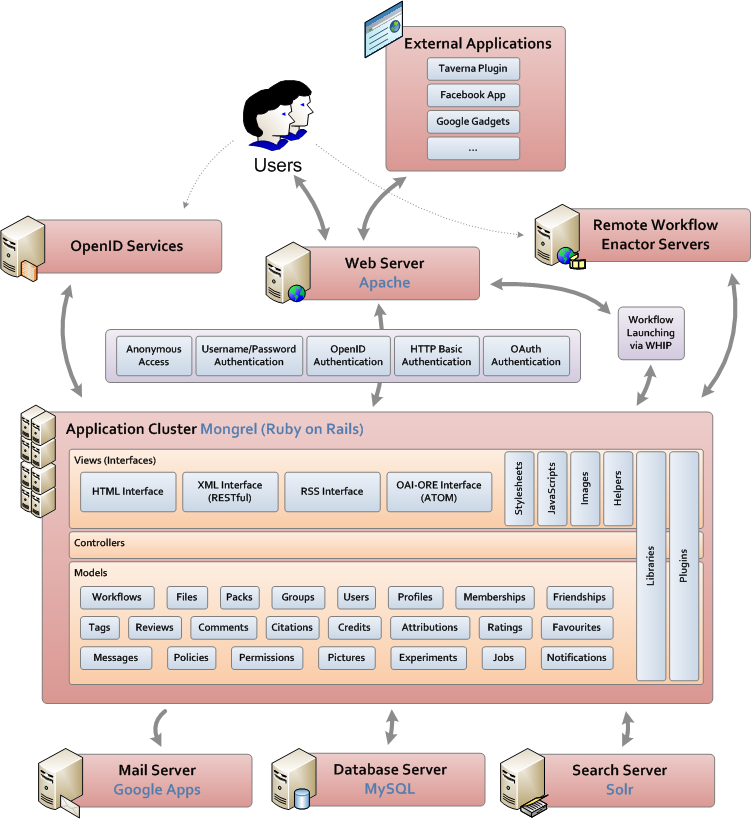
myExperiment was designed according to an interpretation of the Web 2.0 design principles in the context of the virtual research environment [10] and our developer documentation is available on wiki.myexperiment.org. The architecture of one instance of myExperiment is shown below in Figure 2. For ease of use, all the interfaces to myExperiment functionality are accessed via the HTTP protocol. For end users we provide the HTML based Web interface, while external applications can also access the other interfaces, in particular the managed RESTful API. The HTML interface makes minimal assumptions about the capabilities of the browser, optionally using JavaScript and AJAX to improve the interactive experience, while the API enables the construction of Rich Web Applications and mashups.
myExperiment, which is released under the BSD licence, is built in the Ruby on Rails web application framework and follows the Model View Controller abstractions set out in Rails. By keeping with the architectural design of Rails we were able to leverage many of its capabilities to build features for users rapidly. The database server, search server and external workflow enactors are all separate systems with simple interfaces to which the main application connects. Various mechanisms for authentication are provided based on the interfaces used; for example, for end users, authentication can be via external OpenID services. We also provide a SPARQL endpoint for the public myExperiment content, enabling queries of the collection and also the social network. The myExperiment ontology is modularised and conforms with terms from Dublin Core, FOAF, SIOC, OAI-ORE and Creative Commons. This framework enables us to express relationships within and between the research objects held by myExperiment.
Growth and future plans
Since the beta launch in November 2007, myexperiment.org has acquired over 16,000 unique visitors, and 1500 registered users who are sharing over 580 workflows covering a variety of scientific and humanities domains; at the time of writing there are also 133 groups, 164 files and 47 packs. The collection serves both researchers and learners, ranging from self-contained, high value research analysis methods referenced by the journal publications that discuss the results of their use, to training workflows that encode routine best practice scientific analyses or illustrate new techniques for new kinds of research data. Analysis of myexperiment.org usage statistics demonstrates: (i) a rapidly growing community, (ii) extensive use of contributed research objects and (iii) the development of social groups. We are already supporting the Taverna (www.mygrid.org.uk) and Trident (www.microsoft.com/mscorp/tc/trident.mspx) workflow systems and will be adding Kepler (kepler-project.org) and Meandre (seasr.org/meandre), and support for new forms of experiment plans is also being developed.
We have a constant flow of new requirements from our users and our community champions, which we prioritise and work to deliver the requested functionality. As the collection grows we are enhancing the discovery functionality [11] through more advanced search, enabling user-provided controlled vocabularies for smarter discovery, implementing comprehensive analytics for effective recommendations, and developing richer expression of relationships between items. We are also developing more sophisticated interworking with other instances and other repositories, together with added value services such as terminology services, so that users can work seamlessly with the combined collections of research content. We plan to augment our community curation approach to tackle content decay with automated tools to assist users, experts and providers, and thereby investigate the curation of processes as well as data.
myExperiment is part of our vision of the “e-Laboratory” – a set of open tools, services and resources which can be assembled to meet the needs of the researcher, where the utility of the components transcends their immediate application. These include repositories, monitoring and annotation services, provenance capture and analytics, research object management, blogs and wikis – working in silico and in the laboratory [12]. We are evolving towards this through myExperiment and the family of projects around it, such as the BioCatologue project which is providing a curated catalogue of Life Science Web Services (www.biocatalogue.org).
Conclusion
myExperiment is an innovative repository which brings the new artefacts of digital science into the scholarly knowledge cycle. While social extensions to repositories have been discussed before [13], myExperiment is built from the ground up on Web 2.0 principles and is distinct in principle and operation from general repository software: it brings a social infrastructure which facilitates discovery and sharing, and a technical infrastructure which facilitates integration with the research environment and with computational services. It is distinctive among other projects which have focused on bringing data into the scholarly knowledge cycle [14] due to its emphasis on sharing and reuse of process and method. Our vision is that myExperiment users will be able to find the resources they need with ease at point of use, have sophisticated control over privacy and benefit from sharing while they work, disseminate the outcomes of their work and their methods with ease so that they build reputation and facilitate the research of others, and that these items will benefit from curation by their providers, by experts and by the community that is using them.
References
[1] Gil, Y., Deelman, E., Ellisman, M. et al. “Examining the Challenges of Scientific Workflows”. IEEE Computer 40(12): 24-32. 2007.
[2] Goble, C. and De Roure, D. “Curating Scientific Web Services and Workflows”. Educause Review, 43 (5). EDUCAUSE Review, vol. 43, no. 5, September/October 2008.
[3] Goble C, Stevens R, Hull D, Wolstencroft K, Lopez R. “Data curation + process curation=data integration + science”, Brief Bioinform. 2008 Nov;9(6):506-17. Epub 2008 Dec 6. PMID: 19060304
[4] “MySpace for the dudes in lab coats”, New Scientist magazine, issue 2574, page 29. 21 October 2006.
[5] Jim Hendler. “Reinventing Academic Publishing, Part 3”. IEEE Intelligent Systems Jan/Feb 2008, pp. 2-3.
[6] Lincoln Stein. “Towards a cyberinfrastructure for the biological sciences: progress, visions and challenges”. Nat Rev Genet, 9(9):678-688. September 2008.
[7] De Roure, D., Goble, C., Bhagat, J., Cruickshank, D., Goderis, A., Michaelides, D. and Newman, D. “myExperiment: Defining the Social Virtual Research Environment”. In 4th IEEE International Conference on e-Science, 7-12 December 2008, Indianapolis, Indiana, USA.
[8] Lin, Y., Poschen, M., Procter, R., Voss, A., Goble, C., Bhagat, J., De Roure, D., Cruickshank, D. and Rouncefield, M. “Agile Management: Strategies for Developing a Social Networking Site for Scientists”. In 4th International Conference on e-Social Science, 18-20 June 2008, Manchester, UK.
[9] De Roure, D. and Goble, C. “Software Design for Empowering Scientists,” IEEE Software, vol. 26, no. 1, pp. 88-95, January/February 2009.
[10] De Roure, D., Goble, C. and Stevens, R. “The Design and Realisation of the myExperiment Virtual Research Environment for Social Sharing of Workflows”. Future Generation Computer Systems 25, 2009. pp. 561-567.
[11] Goderis, A., Fisher, P., Gibson, A., Tanoh, F., Wolstencroft, K., De Roure, D. and Goble, C. “Benchmarking Workflow Discovery: A Case Study From Bioinformatics”. Concurrency: Practice and Experience. (In Press)
[12] Coles, S. and Carr, L. “Experiences with Repositories & Blogs in Laboratories”. In Third International Conference on Open Repositories 2008, 1-4 April 2008, Southampton, UK.
[13] Ikeda, D. and Inoue, S. “A Sustainable Model based on the Social Network Service to Support the Research Cycle”. .In Third International Conference on Open Repositories 2008, 1-4 April 2008, Southampton, UK.
[14] Coles, S. and Lyon, L. “The eCrystals Federation”. In Third International Conference on Open Repositories 2008, 1-4 April 2008, Southampton, UK.