Research in SENSe
Algorithmic Biology
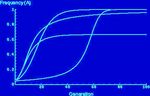
The use of computational modelling and complexity theory to understand the underlying algorithmic principles of biological systems.
Example topics: the major transitions in evolution, the evolution of sex, symbiosis and symbiogenesis, ecosystem selection in biofilms, networks.
Bio-inspired Computing

The development of algorithms and architectures inspired by natural selection and the decentralised organisation of swarms, brains, immune systems, etc.
Example topics: immune system behaviour, coevolutionary algorithms for multiplayer games of imperfect information, decentralised computing architectures, neuromodulation, robust analog circuits.
Biological Self-assembly

The study and manipulation of self-assembling biological molecules and their designed synthetic analogues to enable bottom-up construction of nano/micro-scale devices such as biosensors.
Example topics: lipidic building blocks of cell membranes, lipid polymorphism, semiconductor-tethered liposomes, biotin-avidin interactions, supramolecular complexes, drug delivery.
Complex Networks
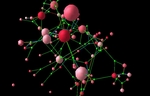
Applying and extending network theory to deal with the large-scale and topologically complex networks found in domains such as biology, computing, geography and knowledge representation.
Example topics: constraints on networks due to spatial layout, integration of multiple interacting networks, network sampling, network structure in markets, ontology networks, social networks.
Ecological & Evolutionary Modelling

The application of modelling techniques developed in artificial intelligence to problems in ecology and evolution.
Example topics: collective building behaviour in insects, the evolution of signalling and communication, social learning behaviour, mimicry, development of sophisticated wireless sensor nodes, the epistemological status of evolutionary simulation models.
Informed Matter

Eliciting the principles of nature's molecular level information processing and applying them to the development of molecular devices and the directed complexification of matter.
Example topics: The use of biological cells in robot control, microfluidic systems for enzymatic computation, computational design of molecular components for information processors, theoretical models of noise-driven behaviour in stochastic genetic and signalling systems.
Some additional information on the research interests of individual group members is also available.


