Geodise Database Toolbox
Introduction
The Geodise Database Toolbox consists of client and server tools which enable distributed users to easily manage, share and reuse their data from within the Matlab environment. Users with no database experience can integrate data management into their applications by calling the archive, query and retrieve functions provided by the toolbox. Any data files or Matlab variables can be stored in the Geodise archive. User defined Matlab structures specify additional descriptive information (metadata), which can be queried to easily locate data of interest. The Geodise Database Toolbox allows you to:
· Manage data from the local Matlab environment or remotely in scripts.
· Store files and variables with customized descriptive metadata.
· Organise related data into datagroups.
· Query over metadata to easily locate required data using functions or a GUI.
· Retrieve data based on logical data identities, no need to remember file locations.
· Share data with other distributed users by granting them access permissions.
There are a separate set of server side tools for the Geodise Database Toolbox. Variables and metadata are stored in an Oracle 9i and 10g database as XML, converted using the XML Toolbox. The Geodise Database Toolbox functions call data management services which utilise Grid, Web Service and database technologies with certificate based authentication and authorisation. The server side tools are not described in any detail in this document.
Tutorial
Getting started
Before using the Geodise Database Toolbox you need to register your details in the database by providing your certificate subject to an administrator, who will then assign you a username. To get your certificate subject call gd_certinfo from the Compute Toolbox.
>> subject = gd_certinfo
subject: C=UK,O=eScience,OU=Southampton,L=SeSC,CN=some user
issuer: C=UK,O=eScience,OU=Authority,CN=CA,E=ca-operator@grid-support.ac.uk
start date: Tue Oct 07 13:00:31 BST 2003
end date: Wed Oct 06 13:00:31 BST 2004
subject =
/C=UK/O=eScience/OU=Southampton/L=SeSC/CN=some user
To setup the Database Toolbox call gd_dbsetup which will create a .geodise directory in your home directory and copy the necessary configuration files into it.
>> gd_dbsetup
You will be prompted for details of your file store host (where gd_archive will store your files). Set hostname to a Globus enabled server you have GridFTP permission on, and set hostdir to an existing directory on that server where files can be stored. These settings will be saved in <home_dir>/.geodise/ClientConfig.xml.
A valid proxy certificate is required to use the Database Toolbox functions, and this can be created using the function gd_createproxy from the Compute Toolbox.
>> gd_createproxy
A GUI will appear and prompt you for your certificate passphrase. Click the ‘Create’ button to generate the proxy certificate. When this is finished click ‘Cancel’ to close the GUI and press 'Enter' at the Matlab prompt.
See the Compute Toolbox Tutorial for more information on certificates and proxy certificates.
Archiving files
To archive a file from the local filesystem, first create a metadata structure containing some information that describes your file. This can be any combination of doubles, strings, arrays, cell arrays, complex numbers and substructures.
>> m.model.name = 'test_design';
>> m.model.params = [1 4.7 5.3];
>> m.product = 25.5431;
Add some standard information (localName, format, comment, version or tree) about the file.
>> m.standard.comment = 'Test design model file';
>> m.standard.version = '1.2.0';
The file can then be archived with the metadata.
>> fileID = gd_archive('C:\file.dat',m)
fileID =
file_dat_c6afa4b4-03cb-49a4-8c4e-008c38aae413
In addition to the optional metadata structure, gd_archive takes a string representing the path and filename of a local file. It stores this file on a remote file store (specified in <user_home>/.geodise/ClientConfig.xml). An ID is returned which is a unique handle that can be used to retrieve the file.
The metadata is stored in a database and can be queried to help you find relevant files. When the file is archived some additional metadata is automatically generated and stored in the standard substructure, regardless of whether user defined metadata was also provided. This consists of localName (the original name of the file), byteSize, format, archiveDate, createDate (when the original file was created/modified) and userID. See gd_query for further information on these fields. You can specify your own overriding values for standard.localName and standard.format if you prefer. You can also include the optional user defined metadata fields comment, version and tree. To help data organisation the tree field can be assigned a hierarchy string, similar to a directory path, e.g. 'myuserID/designs/testmodel'.
Querying file metadata
To query file metadata pass a query string to the gd_query function. A query takes the form 'field = value', where = can be replaced by other comparison operators. More than one query condition can be included in the string using & to join them together. A call to gd_query returns a cell array of structures, one for each matching result.
>> result = gd_query('standard.version=1.2.0 & product>25.4')
result =
[1x1 struct]
>> result{1}
ans =
standard: [1x1 struct]
model: [1x1 struct]
product: 25.5431
gd_display is a convenient way to view your query results.
>> gd_display(result)
*** Content of the structure result{1} (Total structures: 1)
standard.ID: file_dat_c6afa4b4-03cb-49a4-8c4e-008c38aae413
standard.localName: file.dat
standard.byteSize: 24
standard.format: dat
standard.createDate: 2004-09-15 15:25:33
standard.archiveDate: 2004-10-07 11:03:10
standard.userID: jlw
standard.comment: Test design model file
standard.version: 1.2.0
standard.datagroups:
model.name: test_design
model.params:
1.0000 4.7000 5.3000
product: 25.5431
*** No more results. ***
It is possible to select which metadata fields are returned in the query results. This is done by passing a string containing a comma separated list of these fields as the third argument to gd_query. The second argument specifies that we want to query files, but is normally omitted because it is the default.
>> r = gd_query('product>25','file','standard.ID, model.*');
>> gd_display(r)
*** Content of the structure ***
standard.ID: file_dat_c6afa4b4-03cb-49a4-8c4e-008c38aae413
model.name: test_design
model.params:
1.0000 4.7000 5.3000
To search for some text within a metadata value use the 'like' operator together with % to specify any characters, or _ to specify one character.
>> gd_query('standard.comment like %design m_del%');
The * wildcard can be used to represent an anonymous subfield, or any number of subfields if it appears at the beginning.
>> gd_query('*.name = test_design');
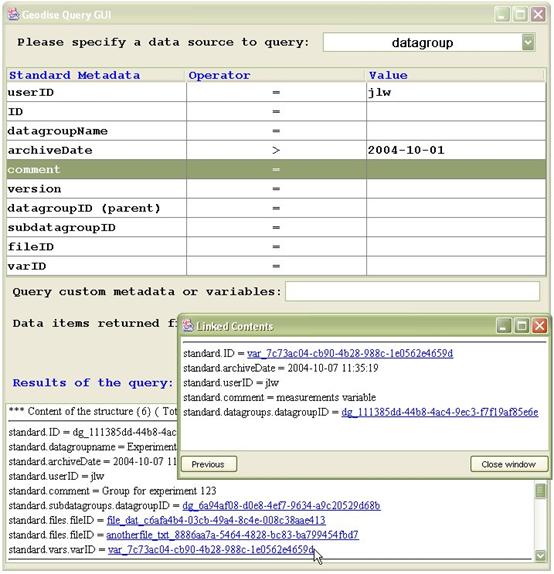
Use gd_query without any input arguments to start the Query Graphical User Interface (GUI), see Figure 2. You can set query conditions for standard metadata by selecting an operator (=, > etc) from the drop down list next to the relevant metadata item and typing in a value. Further query conditions for user defined metadata can be entered in the ‘Query custom metadata or variables’ text field. In the following text field you can enter a comma separated list to specify which metadata items are returned for each matching query result.
Click the ‘Submit Query’ button to run your query. The corresponding gd_query script command is displayed, followed by the results of the query.
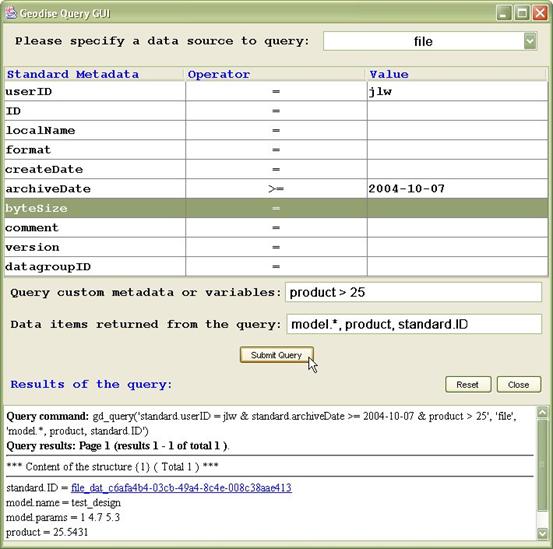
Figure 2 The Query GUI can be used to submit queries and view results.
Hyperlinks are provided in the query results for downloading and browsing data. Figure 3 demonstrates that a file can be downloaded by clicking on its standard.ID hyperlink. In the Save dialog box you can use the default file name value (original name of file) or specify a new file name. Browsing data is further discussed in the Grouping data section.
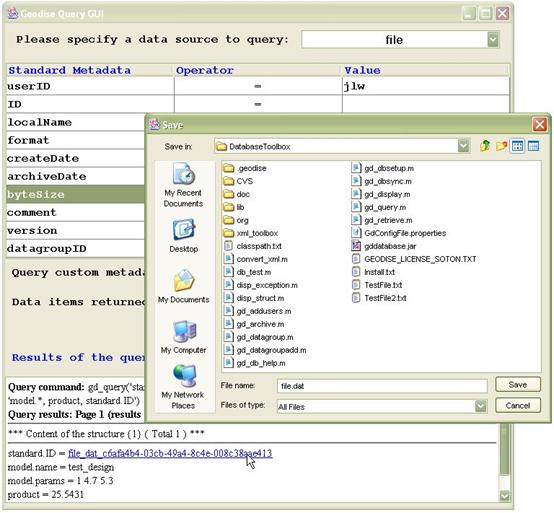
Figure 3 Click on a file's standard.ID link to download that file.
Retrieving files
A file can be retrieved to the local filesystem by specifying its unique ID. This string is returned by gd_archive when the file is archived, and also appears in the metadata query results as standard.ID.
>> ID = result{1}.standard.ID
ID =
file_dat_c6afa4b4-03cb-49a4-8c4e-008c38aae413
The file can be retrieved to a specific file location.
>> gd_retrieve(ID,'C:\filesdir\myfile.dat')
ans =
C:\filesdir\myfile.dat
Alternatively the file can be retrieved to a specified directory (the original file name is used).
>> gd_retrieve(ID,'C:\filesdir')
ans =
C:\filesdir\file.dat
Archiving, querying and retrieving Matlab variables
To archive a variable simply pass it to gd_archive with an optional metadata structure.
>> v.width = 12;
>> v.height = 6;
>> metadata.standard.comment = 'measurements variable';
>> varID = gd_archive(v,metadata);
It is possible to query the contents of an archived structure. Including ‘var’ as the second argument indicates that you want to query the contents of a variable (as opposed to the metadata of the variable).
>> result = gd_query('height=6','var');
>> gd_display(result{1})
*** Content of the structure ***
standard.varID: var_7c73ac04-cb90-4b28-988c-1e0562e4659d
standard.datagroups:
width: 12
height: 6
The contents of the variable are returned along with a small subset of its metadata (standard.varID and standard.datagroups) which may be required for further queries. You can also query a variable's full metadata by including ‘varmeta’ as the second argument.
>> r = gd_query('standard.comment like measure%','varmeta');
>> gd_display(r{1})
*** Content of the structure ***
standard.ID: var_7c73ac04-cb90-4b28-988c-1e0562e4659d
standard.archiveDate: 2004-10-07 11:35:19
standard.userID: jlw
standard.comment: measurements variable
standard.datagroups:
A variable can be retrieved into the local Matlab workspace by specifying its unique ID. This string is returned when the variable is archived (e.g. varID) and also appears in the variable query results as standard.varID and in the metadata query results as standard.ID.
>> v2 = gd_retrieve(varID)
v2 =
width: 12
height: 6
Grouping data
Related data can be logically grouped together using a datagroup as follows:
Specify metadata that applies to the whole group.
>> dgmetadata.standard.comment = 'Group for experiment 123';
Call gd_datagroup to create a datagroup, giving it a name.
>> datagroupID=gd_datagroup('Experiment 123',dgmetadata);
Add archived files or variables to the datagroup.
>> gd_datagroupadd(datagroupID,fileID);
>> gd_datagroupadd(datagroupID,varID);
Archive a new file (with no metadata this time) and add it to the datagroup.
>> gd_archive('C:\anotherfile.txt',[],datagroupID);
The datagroup metadata now contains references to the files and variables it contains. Datagroup metadata can be queried by including ‘datagroup’ as the second argument.
>> r = gd_query('standard.datagroupname=Experiment 123', 'datagroup');
>> gd_display(r)
*** Content of the structure r{1} (Total structures: 1) ***
standard.ID: dg_111385dd-44b8-4ac4-9ec3-f7f19af85e6e
standard.datagroupname: Experiment 123
standard.archiveDate: 2004-10-07 11:42:03
standard.userID: jlw
standard.comment: Group for experiment 123
standard.datagroups:
standard.subdatagroups:
standard.files.fileID: file_dat_c6afa4b4-03cb-49a4-8c4e...
standard.files.fileID: anotherfile_txt_8886aa7a-5464-48...
standard.vars.varID: var_7c73ac04-cb90-4b28-988c-1e0562...
*** No more results. ***
Metadata for the files and variables also contain references to the datagroup(s) that they belong to, with a standard.datagroups.datagroupID field for each datagroup.
Datagroups can be added to other datagroups to create a hierarchy as follows:
>> parentDatagroupID = datagroupID;
>> childDatagroupID = gd_datagroup('child datagroup');
Add the child datagroup (also called a subdatagroup) to the parent datagroup.
>> gd_datagroupadd(parentDatagroupID,childDatagroupID);
Find all the datagroups that are in the parent datagroup.
>> children = gd_query(['standard.datagroups.datagroupID=' parentDatagroupID],'datagroup');
Find all the datagroups that contain the child datagroup.
>> parents = gd_query(['standard.subdatagroups.datagroupID=' childDatagroupID],'datagroup');
Figure 4 Using hyperlinks to browse between related data in the query GUI.
Using the Query GUI you can browse between related datagroups, files and variables by clicking on hyperlinks. In Figure 4 a query on datagroup metadata has been made by selecting datagroup from the drop down list at the top of the window, then specifying the query conditions. The matching datagroup shown in the figure has related subdatagroups, files and variables which are displayed as hyperlinks. Clicking on the standard.vars.varID link brings up a new window containing the metadata for that variable. Clicking on standard.ID in this window will display the contents of the variable itself.
Granting access to data.
The gd_addusers function allows you to grant other users permission to query particular files, variables and datagroups that you own. These users may also retrieve the variables to their local Matlab workspace and the files to their local filesystem (providing they have read permission for the appropriate directory on the Globus file server).
In the following example the user with username ‘bob’ is given access to an archived variable.
>> users = {'bob'};
>> gd_addusers(varID, users);
Access may also be granted as part of the metadata when a file or variable is archived, or when a datagroup is created.
>> m.access.users = {'bob'};
>> gd_archive('C:\file.dat',m);
Further information.
All of these functions have help information which can be viewed by using the help command in Matlab.
>> help gd_display
gd_display Displays the results of a query (a cell of structures), or a single structure.
gd_display(qresults) can be used to display a cell array of structures, e.g. the results of a call to the gd_query function. This is a convenient way of viewing structures to get an overview of their contents.
gd_display(qresults{i}) displays the contents of a structure, e.g. a single result from a query, where i is the index of a structure in the cell array.
Further descriptions and examples for each function are available in the next section of this document.
Copyright © 2007, The Geodise Project, University of Southampton