| University of Southampton > ECS > R4L > Overview | |||||
| Home | Overview | News | Papers and Presentations | Technologies | Background | People | |||||
R4L Project Overview
BackgroundScientific publications, particularly those in the physical science disciplines, invariably report findings that are built upon results gained from experimental data gathering exercises. The processes of gathering the data that underpins a publication can often be very expensive, involved and time consuming, but also information rich and highly valuable to the wider scientific community. In addition, a number of different experiments may be necessary to acquire all the information required to perform a thorough study for publication. The management of data and results from different analyses is currently performed in isolation from each other and as a result comparison, cross reference and identification of common features is time consuming and unreliable and hence seldom performed. Current publication protocols and procedures in the data-based scientific disciplines do not suit the dissemination and sharing of data. A journal article describing the results of scientific work is typically a distillation of experimental data aimed at a wider audience than the immediate peers of the authors. Generally inferences are made only from the most pertinent results, which are reported in a summary format, and journal publication is detached from the production of the experimental data. This renders replication or reuse of the data impossible and results in severe information loss. In addition, access to all the underlying data is either hindered or impossible, again prohibiting further reuse of the data in value added or further studies. A further barrier to unhindered access to scientific data is the ‘licence’ problem, where only researchers in subscribing institutions may access the data held by the publishing body. In the laboratory environment the researcher will perform multiple analyses, as part of a single study, which must be compared and contrasted in order to make deductions. The traditional approach of treating each analysis separately and recording data on different media and formats is both laborious and unreliable, especially when attempting to draw conclusions from comparison between different analyses. Modern computational and scientific instrument technology now allows rapid analyses to be performed, providing the scientist with vast amounts of experimental data, which is becoming increasingly difficult to manage. The emergent field of eScience has the potential to address some of these issues, through the development of Grid-based environments for laboratory experimentation. The EPSRC funded eScience testbed project, CombeChem (http://www.combechem.org) in which a number of project partners were involved, seeks to integrate existing structure and property data sources into an information and knowledge environment. To this end analytical instruments and even synthesis labs were ‘put on the Grid’, enabling the digital output from these operations to be efficiently managed, processed staged and curated using Grid technologies. The technological advances outlined above have caused an explosion of scientific data over the last few years, allowing results to be derived at an unprecedented rate. However, across the scientific domain only a small proportion of the data generated by experimentation appears in, or is referenced by, the published literature. The cause of this shortfall is clearly identifiable as the inability of the traditional publication protocols to take the complete dataset through this process, coupled with an increasing burden placed on the peer review system by the inclusion of just the fraction of the dataset that is conventionally required. This problem may be demonstrated by the current situation with the publication of crystal structures arising from chemical crystallography experiments: A postgraduate student in the 1960’s would have typically investigated around three crystal structures, whilst with the modern technologies available today this may be achieved in a single morning. Despite these advances the publishing protocols for reporting this work are essentially unchanged and in 40 years just 300,000 crystal structures are available in subject specific databases that harvest their content from the published literature. There are around 30million chemical compounds known today and it is estimated that approximately 1.5 million crystal structures have been determined in research laboratories worldwide. Hence less than 20% of the data generated in the crystallographic area is reaching the public domain. As high-throughput technologies, automation and eScience become embedded in scientific working routines the publication bottleneck can only become more severe. The Open Access movement provides a potential solution to this problem through the use of Institutional Repositories (IR) to manage, curate and disseminate academic research output. The scientific papers contained within an IR are either the authors final version of a paper submitted to a journal or the reprint provided by the publisher. The information regarding these papers that is disseminated is purely bibliographic metadata and access, albeit unhindered, is provided only to the content of the paper as it would appear in a journal article. So whilst dissemination of, and access to, academic research articles is greatly facilitated, the paper is still detached from its underlying data. The issue of dissemination and open access to the scientific data underpinning a research publication via an IR has recently been investigated by the JISC funded eBank UK project (http://www.ukoln.ac.uk/projects/ebank-uk), two partners of which are investigators on this proposal. This project has developed a prototype Institutional Data Repository, populated with finalised crystallography results according to a specific and individual schema, in order to investigate mechanisms by which scientific data and experimental results may be disseminated. The Institutional Data Repository makes available metadata regarding the author, affiliation, dataset type and a number of chemical identifiers; the eBank UK project is involved with aggregating this metadata with related studies in the public domain, such as journal articles. Thus the dissemination of scientific data is enabled, but only in parallel to the associated journal article and implicit linking and aggregation between the two is difficult to achieve.
The eBank UK project is concerned with disseminating experimental results (via the development of individual schemas), whereas R4L is about collecting and depositing data and subsequently writing up the results. Currently, eBank supports only a single experimental procedure (crystallography) whereas chemists perform many analyses on compounds (e.g. Spectroscopic: Mass Spectrometry, Nuclear Magnetic Resonance, Ultra-Violet, Infra-Red, Raman; Crystallographic: Single Crystal or powder diffraction; Elemental Analysis; Thermal Analysis and ab-initio quantum mechanical calculations). A goal is to link together, in the Institutional Data Repository environment, all the separate analyses on one particular compound in order to increase the scope of the scientific analysis. It is most likely that only a single repository is involved as a particular institution/investigator is normally responsible for, interested in or co-ordinating the activities of interest on a specific, novel compound. The laboratory repository is a separate entity from the institutional repository not out of architectural necessity, but in order to emphasise a difference in purpose and to ensure the development of appropriate policies (e.g. data storage, access, backup, archiving of raw [proprietary] data using the ATLAS datastore). OverviewIt is possible to make explicit links between the experimental data and article built upon that data if the process is initiated in the laboratory and carried all the way through to the final article. This approach would allow a scientist reading the article access to the underlying data and the experimental processes that generated the data. Thus a scientist would be able to assess and understand the data and reuse it for further value added studies. At the laboratory level the ability for a scientist to pool together various different analyses in a report form and link though to the data provides an extremely powerful form of correlating and cross referencing. The research laboratory is a complex environment and when considered from an eScience perspective consists of a number of administrative domains. The research structure consists of individual researchers, operating as part of a research group and working with other research groups, often in different institutions. Experimental data may be collected on instruments either controlled by the research group or by a different group and often may be obtained by subscribing to an external service. In a scientific study the heterogeneous data produced by these services must be collated and shared with the research group for analysis. The possibilities for aggregating heterogeneous raw experimental data from different sources and experiments, via effective management of the Laboratory Repository, will be explored as part of this proposal and tools developed to enable, manipulate and derive reports for publication purposes.
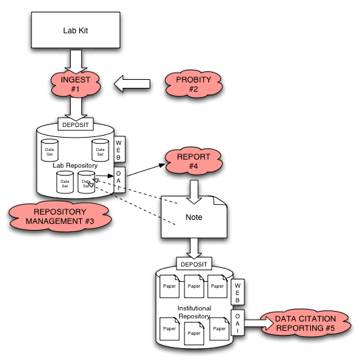
Firstly, data must be captured at the very point of creation in the laboratory. Asides from the raw data acquisition that is the main focus of the experiment, in the case of the ‘smart lab’ subsidiary related data (such as instrument components performance and laboratory environment information by means of sensors etc) would be recorded at source. In many instances this subsidiary information may assist in the interpretation of the raw data, e.g. correlating instrument reading fluctuations with changes in atmospheric conditions such as relative humidity. Instrument manufacturers are interested in the ability to monitor the published performance statistics of an instrument for the purposes of remote fault diagnostics and maintenance, e.g. observing the temperature of a water cooling circuit, where fluctuations could affect the raw data and indicate a fault with the instrument. Of utmost importance is the definition of protocols for capturing the primary raw data in a standard digital format from the instrument as it is being measured. The R4L project will collaborate with instrument manufacturers and a selection of their customers in a requirements capture exercise in order to develop use cases which can evolve standards and protocols for comprehensive raw and subsidiary data capture. Demonstrator systems for testing in this respect would be: Single Crystal and Powder Diffraction, Spectroscopy (Raman, Infra-Red and Mass Spectroscopy) Differential Scanning Calorimetry and aspects of ab-initio quantum mechanics calculations. A semi-automated OAIS INGEST process (#1) would then take data directly from these various types of experimental equipment and deposit it as a data file encapsulated in a dataset in the Laboratory Repository (LR). An automated system has the advantage that it would ensure high quality metadata which conforms to the standards derived from surveys, requirements capture and use cases. The INGEST process would also package equipment operational status data (e.g. instrument calibrations or operational temperature) supporting provenance trails and quality assurance. During the INGEST process (#1) a 'Priority Assertion' service (#2) is invoked, which provides an authenticated and reliable timestamp to certify that the experimental data has been captured at a specific time. This service would provide a legally sound guarantee of priority, which would replace the current need for laboratory notebook signatures and would validate the provenance chain. Although a commercial time-stamping service exists (Surety.com) it does not offer sufficient independence for this application (for a detailed discussion of the proposed service see appendix 1). This is an issue of importance on a global scale. In the USA the ‘first to invent’ is the important factor for claiming a patent, whereas in the rest of the world it is the first to file a claim. Moreover, information from the rest of the world is admissible in US courts and therefore the issue of the ‘first to invent’ affects all. Scientific priority is therefore of interest to all researchers and inventors and it is interesting to note that history suggests that the assigned priority is usually wrong when contested. An enhanced repository management process (#3) is necessary for the R4L, due to the diverse and heterogeneous nature of the different data holdings to be accommodated, arising from different types of experiment. This process manages the various internal data file schemas which document the format of the data files in each data set, corresponding to a particular stage in the experimental procedure. The schemas to be developed for this purpose are quite different from any which represent normal IR composition, or even that used in the eBank dissemination of scientific data project, as established schemas are used to describe a metadata summary of the contents of these files. The schemas to be developed for each experimental process in the R4L context must explicitly describe the format of the data files in each dataset within a holding. High level management would be enabled by rich and thorough generation of metadata. Conventional approaches for describing metadata for dissemination purposes, such as Dublin Core, are not sufficiently exhaustive to describe a scientific dataset. A flexible standard framework that would allow metadata describing these data schema to be used for management and dissemination is the Metadata Encoding and Transmission Standard (METS), which would be thoroughly investigated in this project. A novel approach to the recording or assignment of metadata to digital objects that has been used in conjunction with METS is the Metadata Object Description Schema (MODS), which would also be evaluated for use in this project. This combination would in principle provide a richer element set and greater interoperability, whilst providing extensibility to allow records in other metadata schema to be incorporated into the structure. Having captured all the raw and subsidiary experimental and derived analyses data files, the REPORT process (#4) allows a formal description of the experimental process and relevant data captured from the datasets, for each different experimental analysis performed as part of a scientific study. The description of each experiment process and its relevant data is then combined with a report template (e.g. for a journal submission) to produce a written summary and exposition of the experimental procedure, analysis and conclusions. This report contains statistics, tables and graphs which are automatically generated from the repository's datasets and explicitly stored as RDF triples in the report. The storage of relationships between data files in a repository as RDF triples offers a scalable solution to the issue of storing vast amounts of experimental data. This has been demonstrated by the operation of the CombeChem triple store, which currently contains 60,000 triples and is rising daily with little sign of scalability issues. The resulting report provides a summary of the pertinent data collected from each experiment and not only allows seamless access to ALL the underlying primary and subsidiary data but also enables the researcher to compare and contrast results from the different, but related, experiments. The report is deposited in the Institutional Repository in the normal manner and may be referenced to, submitted for publication or included in a journal article as appropriate. As part of the REPORT process, a standard mechanism must be developed (or adopted) for referring to (i.e. addressing) data items (or collections of data) within a data file inside a data set. Also, a convention must be established for data citation, to allow experimental writeups to explicitly acknowledge the experimental data on which they are built. This is particularly crucial for multi-site collaborations, or for meta-analyses of existing results. Building on this, a data citation service would be established which can track and report the use of experimental data, to establish data value and measures of merit and experimental quality in a way that is analogous to publication quality. In addition the citation tool could be developed to reference inaccurate, outdated, inferior or related datasets or instances in the published literature or public domain. This will be achieved by adapting the Citebase citation analysis service. In the R4L model each research group would therefore operate an Institutional Data Repository for the capture, curation, management, analysis and dissemination of their experimental data. However, often a research group or institution does not possess the instrumentation or facilities for all their analysis requirements and will subscribe to a service e.g. The UK National Crystallography Service (http://www.soton.ac.uk/~xservice). In this instance the Service would be operating a R4L and the user/client would require seamless access to their data. The interaction between a user and a service, through a R4L, is particularly attractive as it would allow the service to retain ownership of the raw data, but make it freely available to a user to use and analyse and generate new results and hence IPR that is independent of the service. The position on IPR and digital rights is currently a prime area of focus in the work that forms the intersection between eBank and CombeChem, documented as the eCrystallography concept. In this area discussions are underway with the managers of the Southampton Institutional Repository, the Centre for Enterprise & Initiative (Southampton University) and we are participating in the current debate on Digital Rights (JISC Consultation workshop, Bristol, 22nd March 2005). A particular component of our attention is the difference between the handling of IPR under different circumstances (i.e. at different stages in the data lifecycle). Firstly the conventional definitions and understanding of IPR are in the expression of ideas, theories and interpretations in articles, which have been well developed in association with traditional modes of publication. However, these definitions do not hold when one wishes to consider ownership and rights in relation to experimental data, which may or may not comprise IPR in the traditional way (for example an author cannot claim IPR for a machine process, which automatically generates data, however non routine interpretations of that data may well contain IPR). The R4L project would seek to continue spearheading the development of approaches to IPR and rights with respect to open publishing of scientific data. Unlike journal articles, no official record of a dataset exists outside the Repository and therefore a data registration process is desirable. Digital Object Identifiers (DOI) are a universally accepted method of registering digital items and will be explored as part of this project. An issue that needs to be resolved is that of the granularity of the DOI reference to the data, i.e. should a dataholding, a dataset, a datafile or a component of a datafile be DOI registered? A final point of interest is that a R4L is designed for use in a research laboratory, yet it is an Institutional archive. This presents issues that are being addressed for conventional institutional archives but would have to be reconsidered in the R4L scenario. These consist of: a) who owns the archive; b) who is responsible for the archive; c) who is the authority governing the archive, which have implications for matters such as policy definition, longevity and preservation.
Aims and ObjectivesThis proposal seeks to apply repository technology to experimental data capture, analysis and reporting processes to enable linking between datasets and articles, and also between related datasets. The primary outcome of this proposed research would be an exemplar system demonstrating the impact of an Institutional Data Repository on the analysis and dissemination of experimental scientific data in a subject that is crucially reliant on such studies. Requirements studies from commercial stakeholders from opposite ends of the scientific experimental process (i.e. equipment manufacturers and learned society publishers) will lead to the development of these pilot services. 1) Consult with scientific equipment manufacturers (as represented by the project partners), data analysis software developers and ‘instruments on the Grid’ eScientists to derive methods and protocols to make raw experimental data available and richly annotated with metadata, as it is generated in the scientific laboratory. 2) Develop an automated OAIS INGEST process which deposits the experimental data and metadata directly into the Laboratory Repository. 3) Establish a pilot ‘Priority Assertion’ service to provide a legally sound guarantee of priority for ‘first to invent’ protection. This service is a scalable, co-operative service designed for an academic context and solving the problems of trust and openness inherent in the alternative (commercial) solutions. A full description of the service is given in Appendix 1, although a cutdown version of this service would be implemented for the scope of this project. 4) Configure a Laboratory Repository to be capable of managing large numbers of heterogeneous scientific datasets. 5) Consult with of an advisory panel composed of scholarly society publishers (as represented by the project partners) to capture requirements for data oriented publishing, including data reference and citation. 6) Build a report editing tool which can integrate repository data into a journal article. Project OutcomesThe following outcomes will be sought from the project: · The majority of scientific data will become freely available, analysable and reusable. · Data sharing will become commonplace, enabling new kinds of science. · Data referencing and citation will allow laboratory output to be assessed alongside journal publications. · Accurate data analyses and report generation. |
|
||||
|
This site is produced and maintained by the R4L project, please contact the
Webmaster for site related comments
|
|||||